Required libraries
── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
✔ ggplot2 3.3.6 ✔ purrr 0.3.4
✔ tibble 3.1.8 ✔ dplyr 1.0.10
✔ tidyr 1.2.0 ✔ stringr 1.5.0
✔ readr 2.1.2 ✔ forcats 0.5.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
Required input files
<- read.delim ("data/VEP/vep_severity.txt" )= read.delim ("data/VEP/pmid29700473_30545852_32461655_dnm_meta_fixMultiVar.txt.gz" )<- read.delim ("data/VEP/pmid29700473_30545852_32461655_dnm_most_severe_gencode.txt.gz" ,comment.char = "#" ,header = FALSE )<- read.delim ("data/VEP/pmid29700473_30545852_32461655_dnm_most_severe_withNovelIsoform.txt.gz" ,comment.char = "#" ,header = FALSE )
Process data and make plot
<- merge (gencode[,c (1 ,2 ,7 )], withNovel[,c (1 ,2 ,7 )], suffixes = c ("_GENCODE" ,"_withNovel" ), by= c ("V1" ,"V2" ))colnames (vep_anno)= c ("Uploaded_variation" ,"Location" ,"Consequence_GENCODE" ,"Consequence_withNovel" )<- vep_anno %>% :: filter (Consequence_withNovel != Consequence_GENCODE)$ severity_gencode= vep_severity$ RANK[match (reassign$ Consequence_GENCODE,vep_severity$ SO.term)]$ severity_withNovel= vep_severity$ RANK[match (reassign$ Consequence_withNovel,vep_severity$ SO.term)]= reassign[reassign$ severity_gencode > reassign$ severity_withNovel,]= merge (reassign,vep_info,by.x= "Uploaded_variation" ,by.y= "varId" )= reassign %>% group_by (Consequence_GENCODE,Consequence_withNovel,Pheno) %>% summarise (N= n ()) %>% spread (Pheno,N,fill = 0 )
`summarise()` has grouped output by 'Consequence_GENCODE',
'Consequence_withNovel'. You can override using the `.groups` argument.
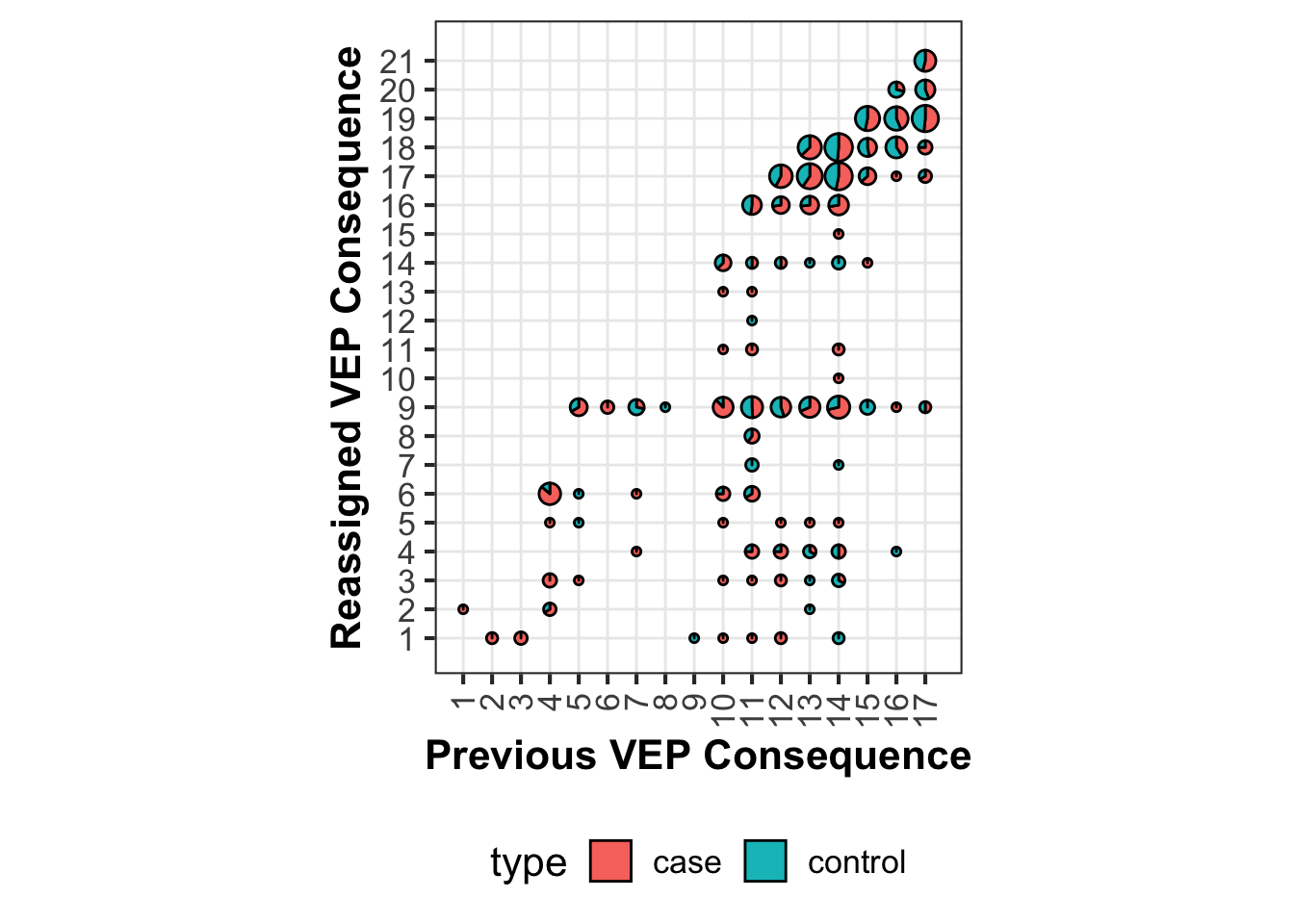
$ severity_gencode= vep_severity$ RANK[match (reassign_sum$ Consequence_GENCODE,vep_severity$ SO.term)]$ severity_withNovel= vep_severity$ RANK[match (reassign_sum$ Consequence_withNovel,vep_severity$ SO.term)]$ Consequence_GENCODE= gsub ("_variant" ,"" ,reassign_sum$ Consequence_GENCODE)$ Consequence_withNovel= gsub ("_variant" ,"" ,reassign_sum$ Consequence_withNovel)$ region= factor (1 : nrow (reassign_sum))$ Consequence_withNovel= reorder (reassign_sum$ Consequence_withNovel, reassign_sum$ severity_withNovel)$ Consequence_GENCODE= reorder (reassign_sum$ Consequence_GENCODE, reassign_sum$ severity_gencode)$ Consequence_GENCODE= as.numeric (reassign_sum$ Consequence_GENCODE)$ Consequence_withNovel= as.numeric (reassign_sum$ Consequence_withNovel)$ total= log (reassign_sum$ case+ reassign_sum$ control+ 1 ,2 ) ^ 0.5 / 6.5 ggplot () + geom_scatterpie (aes (x= Consequence_GENCODE, y= Consequence_withNovel, group= region,r= total),cols= c ("case" ,"control" ),data= reassign_sum) + scale_x_continuous (breaks = seq (1 ,17 ))+ scale_y_continuous (breaks = seq (1 ,21 ))+ coord_fixed ()+ theme_bw (base_size= 16 ) + theme (legend.position = "bottom" ,text = element_text (family = "Arial" ),panel.grid.major = element_line (size= 0.5 ),panel.grid.minor = element_blank (),axis.title = element_text (face = "bold" ),axis.text.x = element_text (angle = 90 , vjust = 0.5 , hjust= 1 )) + ylab ("Reassigned VEP Consequence" ) + xlab ("Previous VEP Consequence" )
Note that geom_scatterpie requires both x and y aesthetics to be numeric, so I converted the VEP consequences into numeric and make the plot. As a result, the corresponding consequences need to be manually added back to the plot.